1. Introduction
Salmonids is a typical cold-water fish with high economic value and promising market prospects. As one of the important economic fish in the world[1], salmonids have been farmed in China for more than 50 years, and the current farmed species include more than 10 species such as Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), silver salmon (Oncorhynchus kisutch), golden trout (Oncorhynchus mykiss, albino phenotype) and Siberian Taimen (Hucho taimen). The scale of farmed salmonids in China is expanding year by year, but the occurrence of viral diseases remains a significant limited factor in aquaculture production[2]. The major viral diseases of salmonids are viral hemorrhagic septicemia (VHS), infectious hematopoietic necrosis (IHN), infectious pancreatic necrosis (IPN) and infection with salmonid alphavirus (ISA)[3]. Infectious hematopoietic necrosis virus (IHNV) and infectious pancreatic necrosis virus (IPNV) are of particular concern for the farmed salmonids in China. Infection caused by each virus has features including rapid transmission, rapid onset, high mortality and difficulty in treatment. They have seriously affected the development of the farmed salmonids industry of China. It is significant that disease causing agents are rapidly identified and differentiated[4]. For this reason, early and accurate nucleic acid detection is crucial and essential in preventing and controlling the spread of salmonids diseases[5, 6].
The biosensors have attracted the researchers due to superior performance in sensitivity, low sample, high throughput and potential for miniaturization[7] and have been widely explored for disease detection, environmental monitoring, food quality monitoring and many other aspects[8-10]. Biosensors consist of two main components: recognition element and transducer. According to the type of the recognition element, biosensors can be divided into nucleic acid sensors, immunosensors, cell sensors, enzyme sensors, etc. It can be categorized into optical, electrochemical, and piezoelectric sensors by the transducer[11]. With the recent development of semiconductors, nanomaterials and microfabrication technologies, semiconductor materials are widely used in biosensing. A variety of novel semiconductor biosensors for pathogen detection with better performance were explored[12]. The nucleic acid biosensors containing semiconductor materials are the most promising tools for early and accurate on-site detection of salmonids viral diseases.
Here, we give a brief description of the current development of salmonids viral diseases detection. And then the present limitations and challenges of early detection for salmonids viral diseases are proposed. Finally, advances of nucleic acid semiconductor biosensors which have potential to apply for on-site early detection of salmonids viral diseases are introduced and the prospects are put forwarded.
2. Salmonids viral diseases detection developments
In 2021, the economic losses caused by diseases in China's aquaculture were about 53.9 billion RMB. Due to the rapid transmission, high mortality and difficulty in treatment of salmonids viruses, the viral diseases outbreaks have resulted in significant economic losses for salmonid fishes. IHN and IPN are the most prevalent two viral diseases of farmed salmonids in China. IHN caused by IHNV is one of the most contagious viral diseases in salmonids that should be reported to the World Organization for Animal Health (WOAH), which can lead to significant mortality rates and huge economic losses on the salmonid farming industry[13-15]. The natural host range of IHNV includes many species of wild and farmed salmon and trout[16, 17]. Mortality caused by IHNV can be substantial, even approaching 100%. IPN is a highly contagious disease that affects salmonids reared in intensive culture systems whose etiological agent is IPNV[18]. It was first isolated in China in 1984[19] and can cause higher than 90% mortality in fry and juvenile trout and salmon and lower mortality in adult fish[20]. Survived adult fish can become asymptomatic carriers of the virus, eventually shedding the virus particles to the environment as a source of horizontal transmission[2, 3]. Early detection and depopulation of the infected fishes are the most common practices to control the spread of IHNV and IPNV[21].
Detection and surveillance on farmed salmonids diseases in China mainly follows the traditional workflow which transport live fish to the testing laboratory (Fig. 1). The first step is to collect samples. Live symptomatic or healthy fishes are caught from salmonids farming sites such as ponds, reservoirs, cages and other aquatic animal breeding sites. The second step is transportation. The samples need to be transported to the testing laboratory within 48 h. The sampled fishes need to be preserved in oxygenated devices to ensure survival and need to be kept at 0–10 °C throughout. Finally, after the samples arrived at the laboratories, pathogen screening and testing are carried out.
The tests of salmonids viruses used in the laboratory mainly are the traditional methods including virological, immunological and nucleic acid test methods. The viral culture needs too much time for isolating and authenticating. Moreover, there is no sensitive cell lines for some viruses of aquatic animals[2, 4]. Enzyme-linked immunosorbent assay (ELISA) is the most typical immunological method, which could detect virus antigen or antibody. But for fish virus, ELISA is not suitable for early detection because virus load is low and antibody is not available in the early stage of fish virus infection[11]. Thereby, the immunological method is hysteretic for fish virus detection. In contrast, the nucleic acid test is a timely and sensitive method since nucleic acid can be detected within 24 h of infection[22]. It can also provide faster diagnosis and multiplexing options for the detection of several viruses at the same time[23]. The reverse transcription-polymerase chain reaction (RT-PCR), RT-quantitative PCR (RT-qPCR) and RT-droplet digital PCR (RT-ddPCR) assays have been developed to qualitative or quantitative detection of IHNV or IPNV[5, 18, 24-32]. Pinheiro et al. developed a multiplex RT-PCR for simultaneous detection of major viruses that infect rainbow trout, including IHNV, IPNV and VHSV[3]. And RT-qPCR is recommended for IHNV detection and surveillance as the validated method by WOAH[5, 15, 26, 33]. Suebsing[34, 35] and Gunimaladevi et al.[36] developed the reverse transcription loop-mediated isothermal amplification (RT-LAMP) to detect IHNV, with sensitivity 10-fold higher than nested PCR. The RT-LAMP methods were also successfully developed to detect the IPNV[20, 34, 37]. Rodriguez et al.[4] evaluated the five serological methods and RT-PCR and found that RT-PCR were the most appropriate and sensitive methods for the routine detection of IPNV from affected fish. Therefore, the nucleic acid test is the most commonly used detection method in the diagnosis of aquaculture viral pathogens.
3. Salmonids viral diseases detection limits and challenges
The current salmonids virus detection and monitoring mode are time-consuming, high cost and limited to laboratory. Firstly, the traditional monitoring mode consumes lengthy time and financial costs during the transportation of samples because aquaculture farms are usually located in lakes, reservoirs and sea areas far from towns and transportation arteries. And the temperature, dissolved oxygen, pH and other conditions of the water during transportation are difficult to control, which are distinct from the living environment of fish and virus, leading to the inaccurate results. This will affect the fish farmers take timely and effective prevention and control measures. In addition, as stated, the widely applied detection method in practice concentrated on the traditional nucleic acid-based methods. However, their detection is too traditional and cumbersome to detect on field. Sophisticated instruments, high power condition, long time and professional staff cause traditional RT-PCR or RT-qPCR non-amenable for on-site and real-time detection[11].
The limitations and problems existing related to the above-mentioned encourage researchers to discover and develop more robust and accurate detection systems for efficient on-site detection and screening[38, 39]. To achieve the goal of early detection and timely control, rapid, low-cost, portable with high sensitivity and specificity, nucleic acid detection methods are highly demanded for on-site detection of virus infection early stage. The biosensors are seen as suitable candidates which have fast response, cost-effectiveness, high sensitivity and selectivity for early detection and surveillance of salmonids viral diseases. Therefore, the nucleic acid test combined with sensing technology are more preferred for accurate and on-site detection. Semiconducting materials make the biosensors more accurate and sensitive due to their advantageous properties such as chemical and mechanical stability, high biocompatibility, unique photoelectric properties and sensitive response[40]. The following section will focus on introducing the developed nucleic acid test-based biosensors which were fabricated by semiconducting materials.
4. Nucleic acid semiconductor biosensors
Semiconductor materials such as silicon (Si), TiO2, quantum dots (QDs) and indium tin oxide (ITO) are widely used in biosensors. Researchers have developed highly sensitive nucleic acid amplification biosensors and amplification-free biosensors based on the properties of semiconductor materials. In addition, microfluidics researches on on-site detection highlights integrated sample-to-detection chip for nucleic acid test assays. They can realize nucleic acid tests implemented on a portable, miniaturized, and integrated device with rapid and sensitive results readout[41, 42]. The integration of microfluidic technology and biosensors can also become a promising tool to shorten detection time and achieve high-throughput detection[43-45].
4.1 Nucleic acid amplification biosensors
In combination with classical PCR amplification and isothermal amplification technology such as LAMP, helicase dependent amplification (HDA) which are more suitable for on-site detection[46], many biosensors based on nucleic acid amplification were developed.
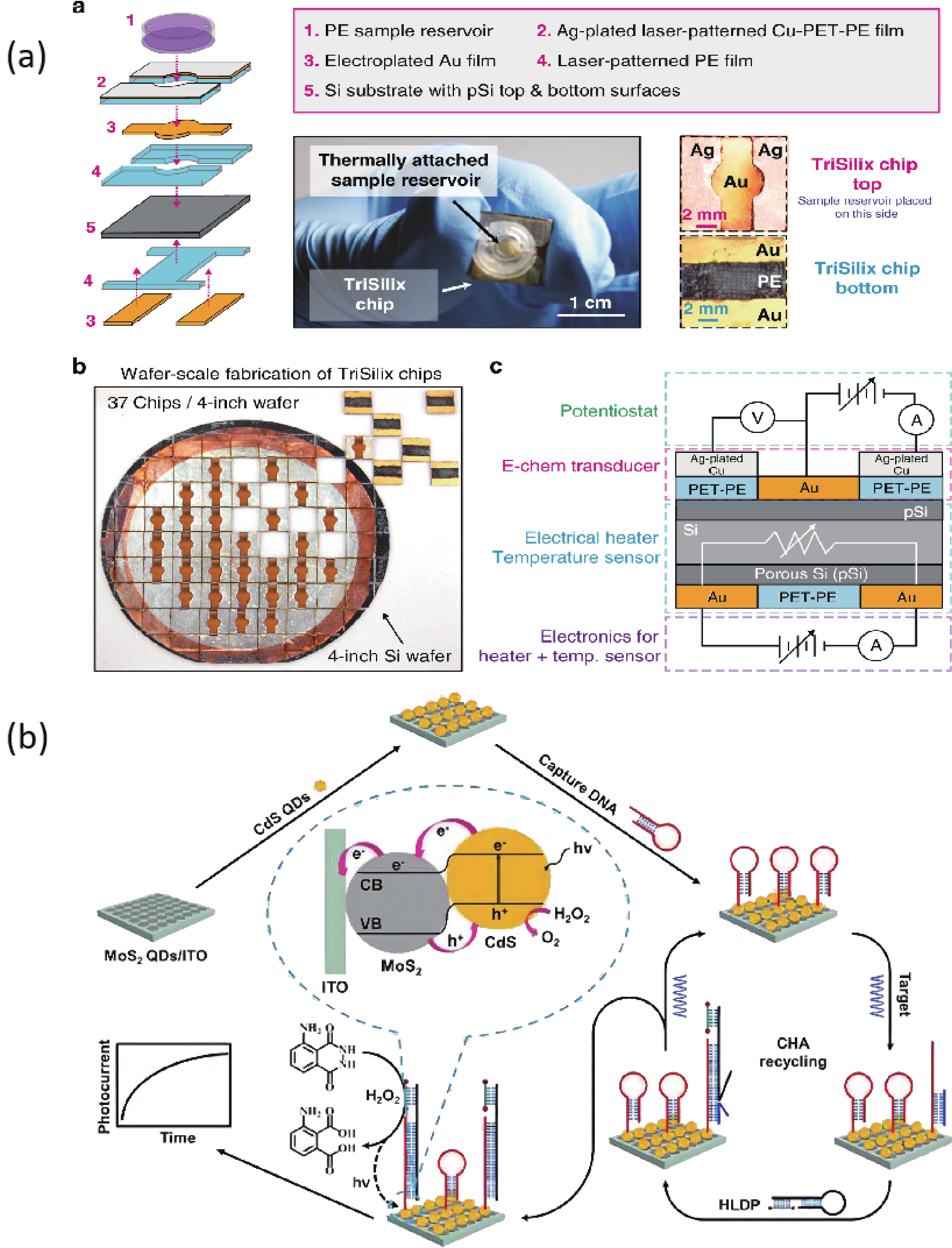
Si is the most commonly used material in large-scale semiconductor biosensors manufacturing, which can greatly reduce the fabricating costs. A portable and battery-powered PCR device which integrated a PCR thermal control system, a PCR reaction chip and the end-point fluorescence detection system was developed by Jie et al. The disposable reaction chips were fabricated with a silicon substrate due to its excellent thermal conductivity, silicone rubber and a quartz plate. The PCR thermal control system was developed using Al thin-film heaters and RTDs to provide fast and precise temperature conditions with an average heating rate of 32 °C/s and an average cooling rate of 7.5 °C/s. The time consumption of the 40 cycles of PCR was less than 1 h[47]. Sun et al. realized to detect five equine respiratory diseases pathogens simultaneously on a silicon microfluidic chip using digital LAMP. Silicon chips are highly stable, have no auto-fluorescence and can be manufactured easily and efficiently. The chips contained ten parallel flow channels with a volume of 1 μL each. The chip surface was thermally oxidized to grow a 200-nm layer of SiO2 that used to reduce the potential for bare silicon to inhibit the amplification process. The detection took place within 30 min with LOD of 18 copies per reaction[48]. Powell and colleagues developed rapid and sensitive nucleic acid amplification tests for HCV, HIV, Zika, HPV16 and HPV18 on the same silicon microchip platform integrated nucleic acid extraction and PCR. These on-chip assays were performed in under 25 min and the efficiency was similar to the assay performed on a bench scale real-time PCR instrument[49]. Nunez-Bajo and co-workers reported a disposable, ultra-low-cost integrated silicon-based on-site transducer (TriSilix) for pathogen nucleic acid amplification and real-time detection. Si was selected as the substrate material to make itself can be used both as an electrical heater and temperature sensor without adding any additional components for temperature transduction. Both isothermal and cyclic nucleic acid amplification can be performed and it integrated temperature control and DNA detection on the same device, which was label-free, requires minimal sample handling. And the TriSilix chips did not need cleanroom for fabrication and can be produced in a standard wet-lab with easily accessible laboratory equipment worldwide. TriSilix was an ideally low-cost, high-performance, on-site nucleic acid detection technology[50] (Fig. 2).
ITO is a conductive and transparent oxide that has advantages of high stability, ease of combination with microfabrication processes, which is used as an electrode material for biosensors[51]. Barreda-Garcia and co-workers utilized the advantages of ITO to develop an optimized and nucleic acid amplification-based biosensor for detecting salmonella. They designed an electrochemical DNA sensor based on a sandwich hybridization assay on the surface of ITO electrodes, which realizes monitoring both PCR and HDA amplicons. ITO was used as a thin film heating element and the reaction surface[52, 53].
Molybdenum disulfide (MoS2) is a typical transition-metal dichalcogenide semiconductor which has unique optical, electronic and catalytic properties. Zang and coauthors developed a CdS/MoS2 heterojunction-based photoelectrochemical DNA biosensor. The CdS/MoS2 photocathode was produced by the stepwise assembly of MoS2 and CdS QDs on ITO. The dual hemin-labeled DNA probe was introduced on high-efficiency CdS/MoS2 electrode via catalytic hairpin assembly (CHA) cycling reaction. This ultrasensitive biosensor achieved a detection limit of 0.39 fM[54] (Fig. 2).
4.2 Nucleic acid amplification-free biosensors
The principle of nucleic acid amplification-free biosensors depends on the hybridization through spontaneous hydrogen bonding between the target nucleic acid and its complementary strand[55]. The biosensors are usually utilized by immobilizing the single-stranded DNA onto a suitable surface. Au nanorods, CdTe QDs and metallic oxide and so on are growing frequently used to fabricate sensor surface as a promising semiconductor material.
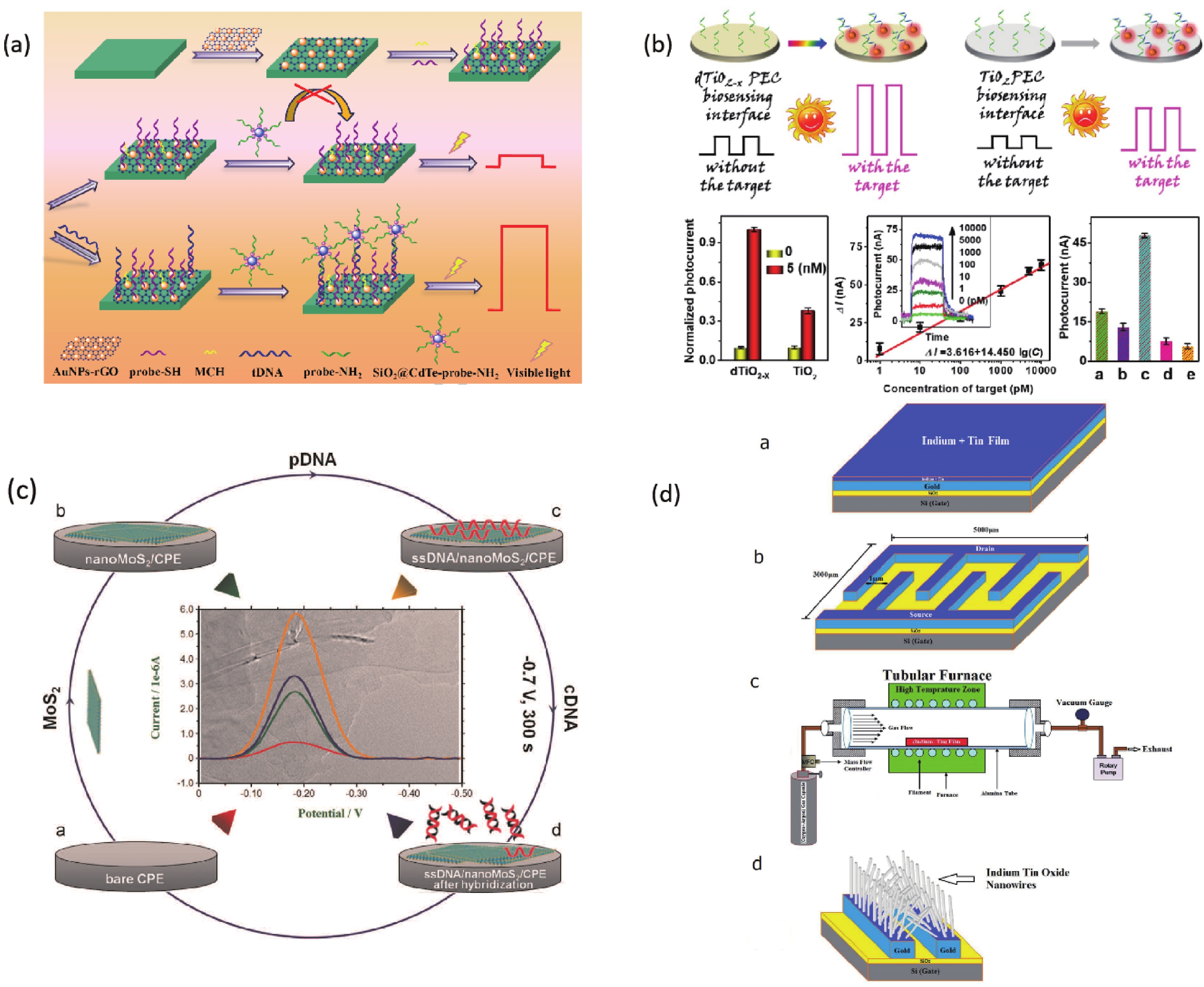
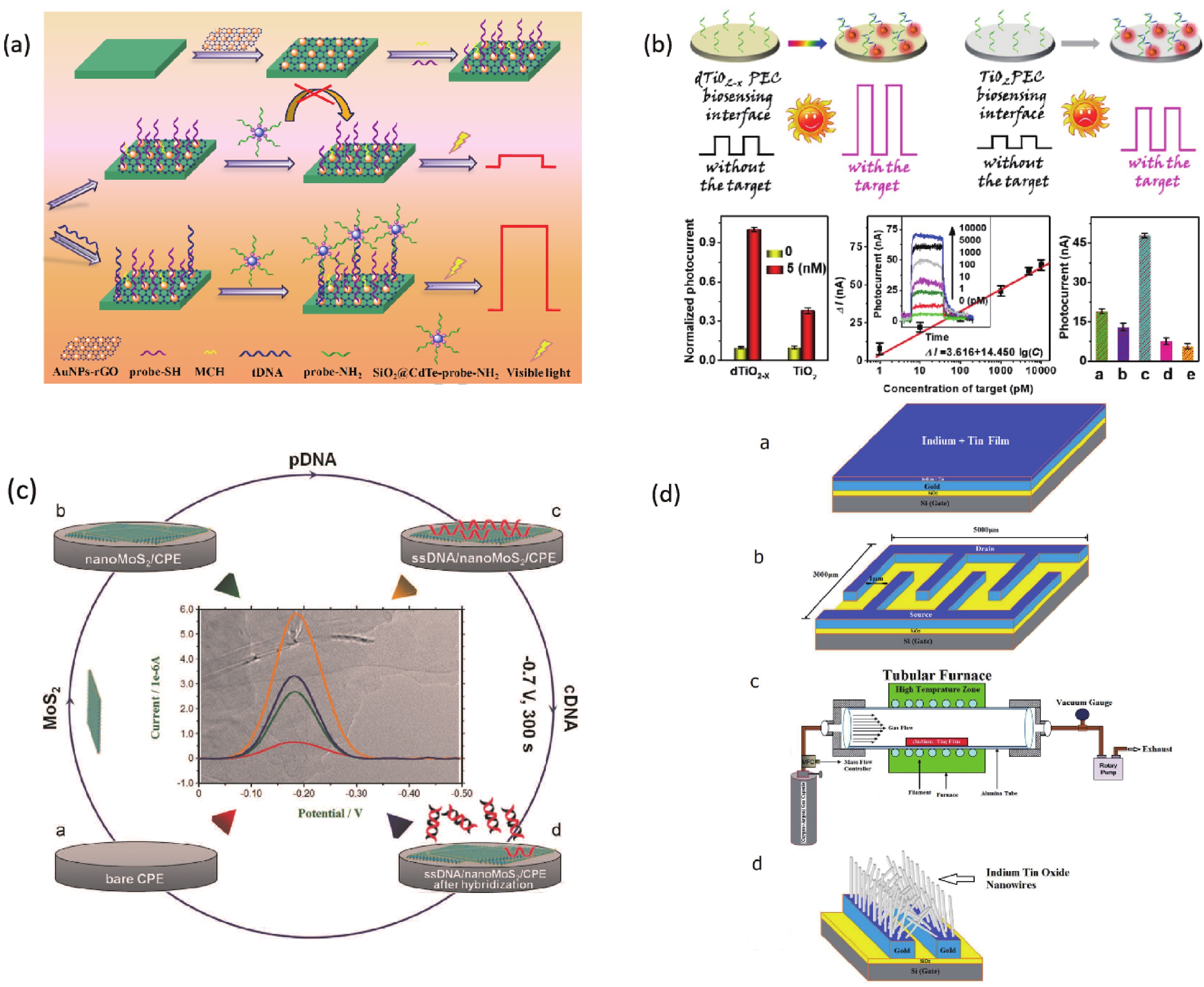
A biosensor for the Cauliflower Mosaic Virus 35S (CaMV35S) promoter detection was reported to distinguish transgenic soybean from non-transgenic samples. The Au nanoparticles-reduced graphene oxide acted as a nanocarrier to immobilize the thiol-functional probe, and the SiO2@CdTe QDs core-shell nanoparticles tagged with the amino-functional probe acted as signal indicators, respectively. In the presence of target DNA of CaMV35S, the binding of tDNA with two probes through the high specific DNA hybridization led to the fabrication of sandwich structure, and thus the high loading of the signal indicators SiO2@CdTe QDs at the electrode surface, which increased the PEC signal. The increased signal depended on the concentration of tDNA and the low detection limit of 0.05 pM was obtained[56] (Fig. 3). Zhang et al. found that monolayer MoS2 has high fluorescence quenching efficiency and shows different affinity towards the single-stranded DNA and double stranded DNA. Therefore, a thin-layer MoS2 nanosheets based and label-free DNA sensor was constructed, with a detection limit of 1.9 × 10−17 M[58, 59] (Fig. 3).
TiO2 is a classical n-type semiconductor, its production method is simple. The raw materials are cheap and easily available. More importantly, it has good biocompatibility, excellent photostability, fast charge conduction rate and excellent photoelectric chemical activity. Many biosensors have been achieved high-performance based on TiO2[60]. A photoelectrochemical DNA sensor based on FTO/TiO2/Au electrode was successfully developed by Liu and coworkers. Combining the small band gap of CdSe QDs with the large band gap of TiO2, the photoelectric conversion efficiency was enhanced. The limit of detection was 3 fM[61]. Tang’s research group modified the electrode surface with the defect-engineered TiO2-x nanorods as photoelectric materials. A photoelectrochemical biosensor platform was constructed for sensitive analysis of target DNA with the surface plasmon resonance enhancement technology of Au nanoparticles. The introduction of Au NPs can not only further enhance the photoelectric signal response but also improve the analytical performance of the sensor on the target DNA. This signaling format combined with exonuclease III-assisted target recycling amplification shows excellent analytical performance[57] (Fig. 3).
Chen and co-workers developed a nanostructured impedance biosensor to detect hepatitis B virus (HBV) DNA. The anodic aluminum oxide membrane having a uniform nano hemisphere array was selected as the substrate of the sensing electrode. A 30-nm gold film was sputtered onto the Al2O3 layer surface as the electrode, followed by electrochemical deposition of Au nanoparticles on each hemisphere surface. Target HBV DNA was captured by hybridization with the specific probe which immobilized on the nanostructured electrode. The LOD was measured to be 111 copy/mL[62]. Zhu et al. developed a DNA sensor to detect HBV with a Cu2O hollow microsphere that can be rapidly synthesized at room temperature. The device has a sensitive detection response and good selectivity for hybridization detection with a detection limit of 1.0 × 10−10 M. This is mainly due to the high specific surface area of Cu2O nanoparticles and the high probe affinity of the particle surface to ssDNA[63]. Shariati et al. reported an ITO nanowires based DNA biosensor compatible with metal-oxide-semiconductor (CMOS) technology to detect HBV. The immobilization of thiolated HBV single-strand DNA is supported with the employment of ITO nanowires reaching the detection limit of 1 fM[64] (Fig. 3). Shariati’s group also developed the HBV detection biosensor whose heterojunction optical electrodes based on WO3/In2O3 nanowires doped with ITO with high sensitivity and stability[65]. Manzano and co-workers reported a label-free user-friendly electrochemical DNA biosensor for hepatitis A virus (HAV). The hybridization reaction occurs on the disposable electrode made of gold functionalized with specific capture probes. The results are monitored through the oxidative peak potential by using cyclic voltammetry[66].
5. Perspectives and conclusions
Considerable efforts of researches demonstrate the utility of robust nucleic acid tests biosensors with semiconductor for development of a flexible nucleic acid diagnostic platform. With the advantages of semiconductor materials, these biosensors have their own characteristics in rapidity, low cost, high sensitivity and specificity. Accurate and sensitive bioanalysis of nucleic acids is crucial for earlier detection of salmonids viral diseases. Currently, there are no practical applications of semiconductor-based biosensors for salmonids virus detection, but these biosensors have great potential application prospects for the diagnosis of salmonids virus infections in the field. Their portable, rapid and sensitive prosperities could meet the needs of salmonids diseases detection and achieve the goal of early detection of salmonids virus infections.
In spite of wide application prospects of biosensors in salmonids virus detection, there are still some challenges in on-site virus detection and surveillance in salmonids aquaculture. The development trends of on-site virus detection biosensors are mainly in these aspects: (1) Miniaturization and integration of the detection system. It will accord with the diagnostic needs of point-of-care (POC) in resource limited settings such as fish farms. (2) High availability and low cost. Fabricating biosensors can use the lower-cost and easily available semiconductor materials. (3) Multiplexed detection. This is highly desirable in early salmonids virus detection because virus has genetic variability such as IPNV. (4) Application effect evaluation. All biosensors need to be applied in the practical environment to evaluate their detection effect and performance. These goals will encourage researchers to make great progress in more accurate, portable, ultracompact reliable and smart on-site virus detection[67, 68].
Acknowledgements
The authors thank anonymous reviewers for their constructive comments on the resentation of this article. This research was supported by the National Key Research and Development Program of China (2022YFC2601304) and National Key Research and Development Program of China (2022YFC2602100)





 DownLoad:
DownLoad:

 DownLoad:
DownLoad:











 Xiaofei Liu:got her Master's Degree on Veterinary in 2014 at Jilin University. Her research interests include detection and monitoring technologies for animal diseases, animal disease risk analysis and genetically modified animals detection
Xiaofei Liu:got her Master's Degree on Veterinary in 2014 at Jilin University. Her research interests include detection and monitoring technologies for animal diseases, animal disease risk analysis and genetically modified animals detection Shaoqiang Wu:got his Doctor's Degree on Veterinary Parasitology in 2003 at China Agricultural University. His research interests include precision detection technology for biosecurity risk, on-site detection techniques for shellfish parasites and aquatic animal diseases
Shaoqiang Wu:got his Doctor's Degree on Veterinary Parasitology in 2003 at China Agricultural University. His research interests include precision detection technology for biosecurity risk, on-site detection techniques for shellfish parasites and aquatic animal diseases Xiangmei Lin:got her Doctor's Degree on Veterinary Pathology in 1998 at Nanjing Agricultural University. Her research interests include detection and monitoring technologies for animal diseases, zoonotic diseases, foreign animal diseases and monitoring technologies of genetically modified animals detection
Xiangmei Lin:got her Doctor's Degree on Veterinary Pathology in 1998 at Nanjing Agricultural University. Her research interests include detection and monitoring technologies for animal diseases, zoonotic diseases, foreign animal diseases and monitoring technologies of genetically modified animals detection
 DownLoad:
DownLoad:






