| Citation: |
Wang Xi, Delphine Delacour, Benoit Ladoux. Designer substrates and devices for mechanobiology study[J]. Journal of Semiconductors, 2020, 41(4): 041607. doi: 10.1088/1674-4926/41/4/041607
****
W Xi, D Delacour, B Ladoux, Designer substrates and devices for mechanobiology study[J]. J. Semicond., 2020, 41(4): 041607. doi: 10.1088/1674-4926/41/4/041607.
|
Designer substrates and devices for mechanobiology study
DOI: 10.1088/1674-4926/41/4/041607
More Information
-
Abstract
Both biological and engineering approaches have contributed significantly to the recent advance in the field of mechanobiology. Collaborating with biologists, bio-engineers and materials scientists have employed the techniques stemming from the conventional semiconductor industry to rebuild cellular milieus that mimic critical aspects of in vivo conditions and elicit cell/tissue responses in vitro. Such reductionist approaches have help to unveil important mechanosensing mechanism in both cellular and tissue level, including stem cell differentiation and proliferation, tissue expansion, wound healing, and cancer metastasis. In this mini-review, we discuss various microfabrication methods that have been applied to generate specific properties and functions of designer substrates/devices, which disclose cell-microenvironment interactions and the underlying biological mechanisms. In brief, we emphasize on the studies of cell/tissue mechanical responses to substrate adhesiveness, stiffness, topography, and shear flow. Moreover, we comment on the new concepts of measurement and paradigms for investigations of biological mechanotransductions that are yet to emerge due to on-going interdisciplinary efforts in the fields of mechanobiology and microengineering. -
References
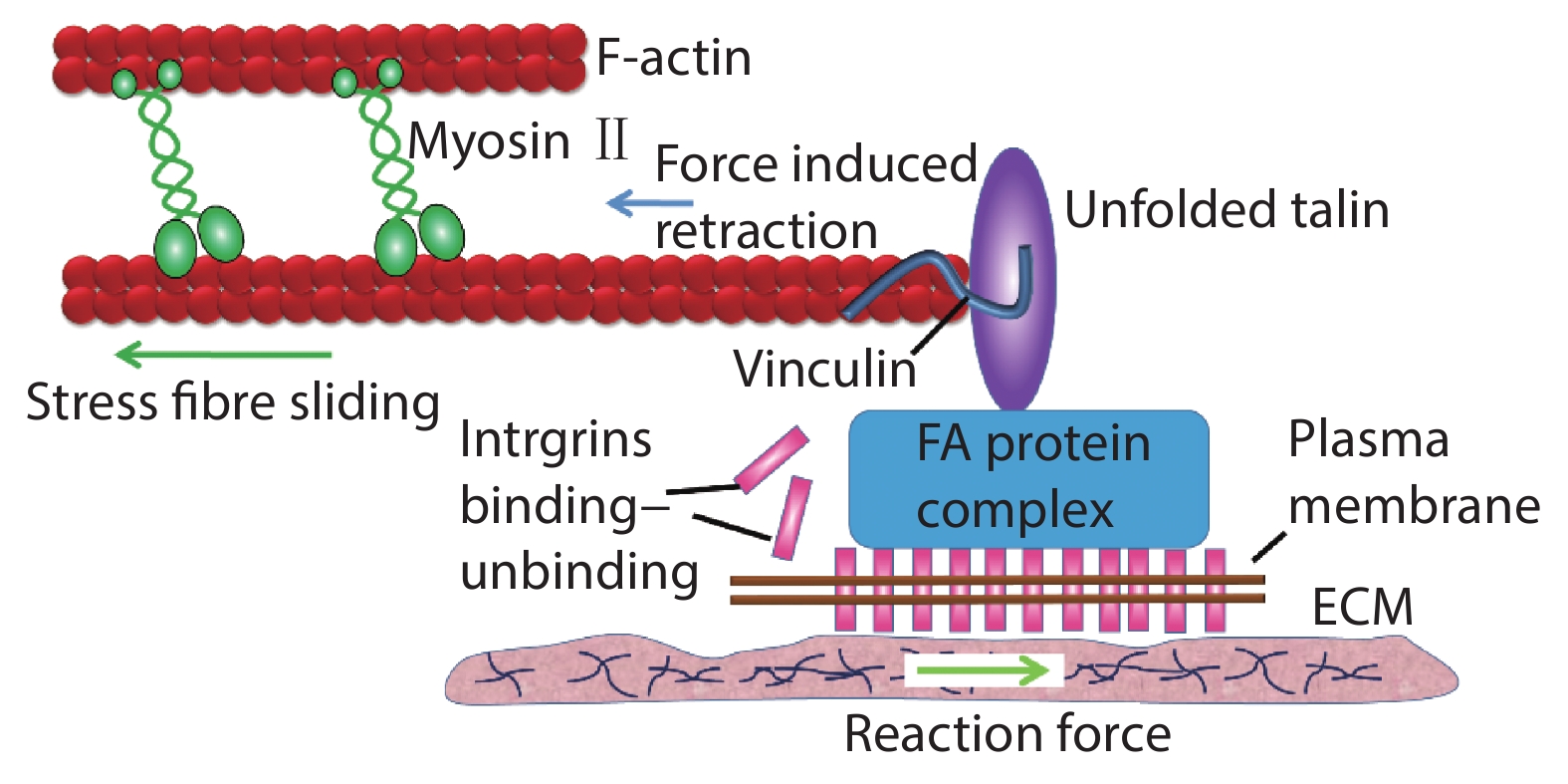
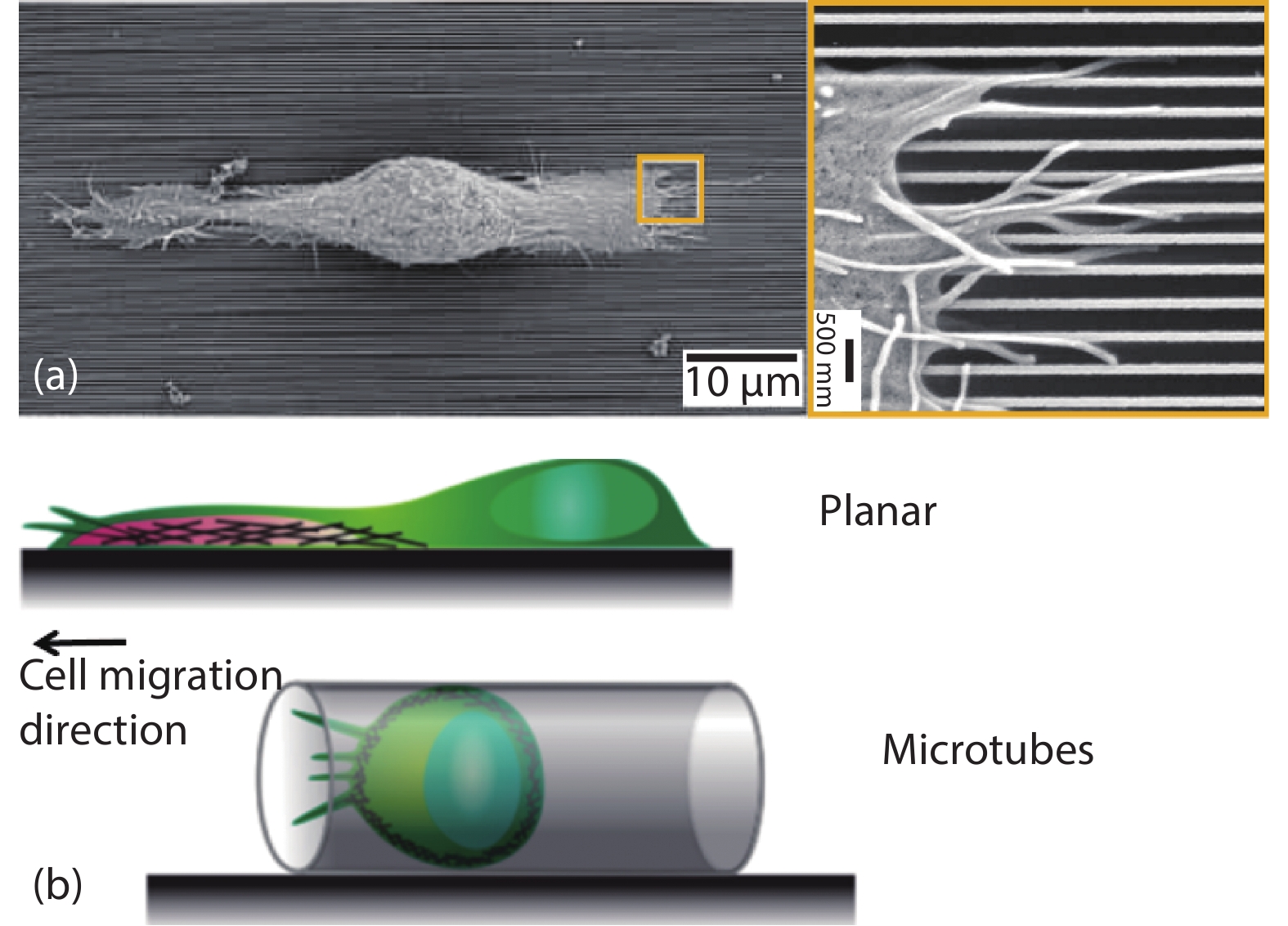
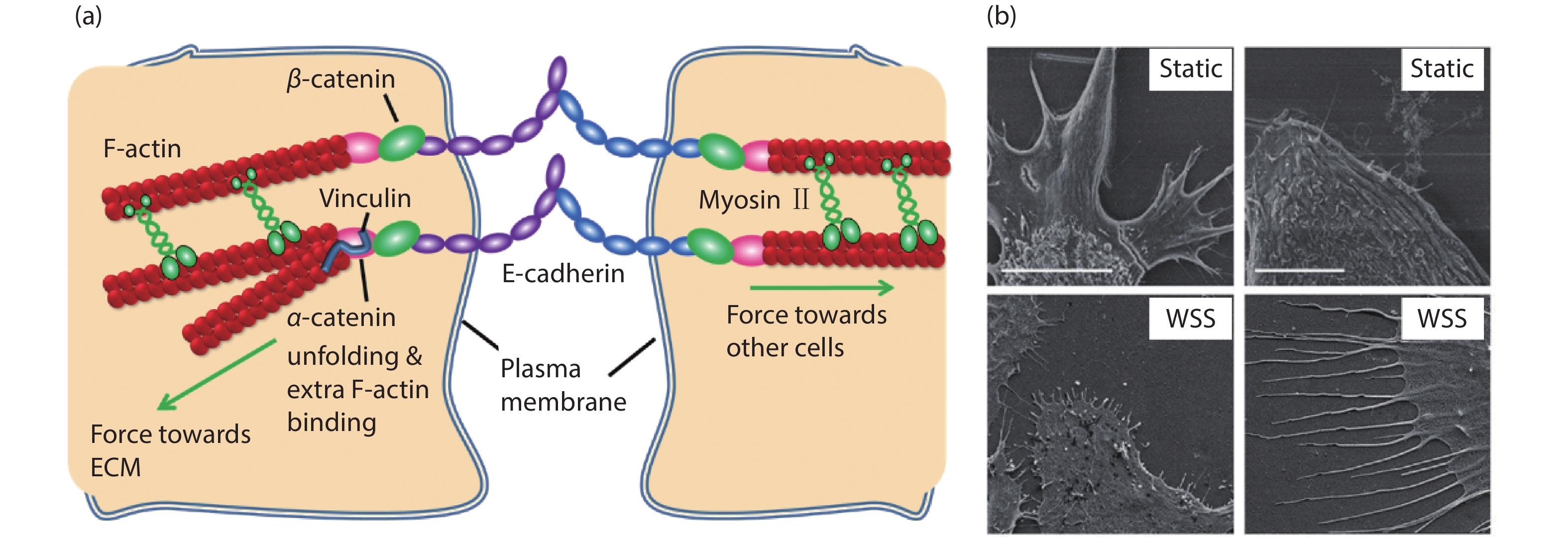
[1] Xi W, Saw T B, Delacour D, et al. Material approaches to active tissue mechanics. Nat Rev Mater, 2019, 4(1), 23 doi: 10.1038/s41578-018-0066-z[2] Prost J, Jülicher F, Joanny J F. Active gel physics. Nat Phys, 2015, 11(2), 111 doi: 10.1038/nphys3224[3] Marchetti M C, Joanny J F, Ramaswamy S, et al. Hydrodynamics of soft active matter. Rev Mod Phys, 2013, 85(3), 1143 doi: 10.1103/RevModPhys.85.1143[4] Needleman D, Dogic Z. Active matter at the interface between materials science and cell biology. Nat Rev Mater, 2017, 2, 17048 doi: 10.1038/natrevmats.2017.48[5] Grashoff C, Hoffman B D, Brenner M D, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature, 2010, 466(7303), 263 doi: 10.1038/nature09198[6] Giannone G, Dubin-Thaler B J, Döbereiner H G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell, 2004, 116(3), 431 doi: 10.1016/S0092-8674(04)00058-3[7] Ray A, Lee O, Win Z, et al. Anisotropic forces from spatially constrained focal adhesions mediate contact guidance directed cell migration. Nat Commun, 2017, 8, 14923 doi: 10.1038/ncomms14923[8] Jiang X, Bruzewicz D A, Wong A P, et al. Directing cell migration with asymmetric micropatterns. Proc Natl Acad Sci USA, 2005, 102(4), 975 doi: 10.1073/pnas.0408954102[9] Chen B, Kumar G, Co C C, et al. Geometric control of cell migration. Sci Rep, 2013, 3(1), 1 doi: 10.1038/srep02827[10] Johnson H E, King S J, Asokan S B, et al. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J Cell Biol, 2015, 208(4), 443 doi: 10.1083/jcb.201406102[11] Engler A J, Sen S, Sweeney H L, et al. Matrix elasticity directs stem cell lineage specification. Cell, 2006, 126(4), 677 doi: 10.1016/j.cell.2006.06.044[12] Naganathan S R, Middelkoop T C, Fürthauer S, et al. Actomyosin-driven left-right asymmetry: from molecular torques to chiral self organization. Curr Opin Cell Biol, 2016, 38, 24 doi: 10.1016/j.ceb.2016.01.004[13] Tee Y H, Shemesh T, Thiagarajan V, et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol, 2015, 17(4), 445 doi: 10.1038/ncb3137[14] Gupta M, Sarangi B R, Deschamps J, et al. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat Commun, 2015, 6(1), 1 doi: 10.1038/ncomms8525[15] Trichet L, Le Digabel J, Hawkins R J, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA, 2012, 109(18), 6933 doi: 10.1073/pnas.1117810109[16] Zemel A, Rehfeldt F, Brown A E, et al. Optimal matrix rigidity for stress-fibre polarization in stem cells. Nat Phys, 2010, 6(6), 468 doi: 10.1038/nphys1613[17] Levina E M, Domnina L V, Rovensky Y A, et al. Cylindrical substratum induces different patterns of actin microfilament bundles in nontransformed and in ras-transformed epitheliocytes. Exp Cell Res, 1996, 229(1), 159 doi: 10.1006/excr.1996.0354[18] Svitkina T M, Rovensky Y A, Bershadsky A D, et al. Transverse pattern of microfilament bundles induced in epitheliocytes by cylindrical substrata. J Cell Sci, 1995, 108(2), 735 doi: 10.1083/jcb.128.4.699[19] Sun B, Xie K, Chen T H, et al. Preferred cell alignment along concave microgrooves. RSC Adv, 2017, 7(11), 6788 doi: 10.1039/C6RA26545F[20] Biton Y Y, Safran S A. The cellular response to curvature-induced stress. Phys Biol, 2009, 6(4), 046010 doi: 10.1088/1478-3975/6/4/046010[21] Bade N D, Kamien R D, Assoian R K, et al. Curvature and Rho activation differentially control the alignment of cells and stress fibers. Sci Adv, 2017, 3(9), e1700150 doi: 10.1126/sciadv.1700150[22] De R, Zemel A, Safran S A. Dynamics of cell orientation. Nat Phys, 2007, 3(9), 655 doi: 10.1038/nphys680[23] Livne A, Bouchbinder E, Geiger B. Cell reorientation under cyclic stretching. Nat Commun, 2014, 5, 3938 doi: 10.1038/ncomms4938[24] Sidhaye V K, Schweitzer K S, Caterina M J, et al. Shear stress regulates aquaporin-5 and airway epithelial barrier function. Proc Natl Acad Sci USA, 2008, 105(9), 3345 doi: 10.1073/pnas.0712287105[25] Ladoux B, Mège R M. Mechanobiology of collective cell behaviours. Nat Rev Molecul Cell Biol, 2017, 18(12), 743 doi: 10.1038/nrm.2017.98[26] Wong S, Guo W H, Wang Y L. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc Natl Acad Sci USA, 2014, 111(48), 17176 doi: 10.1073/pnas.1412285111[27] Saez A, Ghibaudo M, Buguin A, et al. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc Natl Acad Sci USA, 2007, 104(20), 8281 doi: 10.1073/pnas.0702259104[28] Wang X, Li S, Yan C, et al. Fabrication of RGD micro/nanopattern and corresponding study of stem cell differentiation. Nano Lett, 2015, 15(3), 1457 doi: 10.1021/nl5049862[29] Saw T B, Doostmohammadi A, Nier V, et al. Topological defects in epithelia govern cell death and extrusion. Nature, 2017, 544(7649), 212 doi: 10.1038/nature21718[30] Salomon J, Gaston C, Magescas J, et al. Contractile forces at tricellular contacts modulate epithelial organization and monolayer integrity. Nat Commun, 2017, 8, 13998 doi: 10.1038/ncomms13998[31] Wang Y, Gunasekara D B, Reed M I, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials, 2017, 128, 44 doi: 10.1016/j.biomaterials.2017.03.005[32] Stroka K M, Jiang H, Chen S H, et al. Water permeation drives tumor cell migration in confined microenvironments. Cell, 2014, 157(3), 611 doi: 10.1016/j.cell.2014.02.052[33] Raab M, Gentili M, de Belly H, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science, 2016, 352(6283), 359 doi: 10.1126/science.aad7611[34] Denais C M, Gilbert R M, Isermann P, et al. Nuclear envelope rupture and repair during cancer cell migration. Science, 2016, 352(6283), 353 doi: 10.1126/science.aad7297[35] Moreau H D, Blanch-Mercader C, Attia R, et al. Macropinocytosis overcomes directional bias in dendritic cells due to hydraulic resistance and facilitates space exploration. Develop Cell, 2019, 49(2), 171 doi: 10.1016/j.devcel.2019.03.024[36] Xi W, Schmidt C K, Sanchez S, et al. Rolled-up functionalized nanomembranes as three-dimensional cavities for single cell studies. Nano Lett, 2014, 14(8), 4197 doi: 10.1021/nl4042565[37] Xi W, Schmidt C K, Sanchez S, et al. Molecular insights into division of single human cancer cells in on-chip transparent microtubes. ACS Nano, 2016, 10(6), 5835 doi: 10.1021/acsnano.6b00461[38] Koch B, Meyer A K, Helbig L, et al. Dimensionality of rolled-up nanomembranes controls neural stem cell migration mechanism. Nano Lett, 2015, 15(8), 5530 doi: 10.1021/acs.nanolett.5b02099[39] Maquart F X, Monboisse J C. Extracellular matrix and wound healing. Pathol Biol, 2014, 62(2), 91 doi: 10.1016/j.patbio.2014.02.007[40] Kobayashi H, Enomoto A, Woods S L, et al. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol, 2019, 16(5), 282 doi: 10.1038/s41575-019-0115-0[41] Kleinman H K, Philp D, Hoffman M P. Role of the extracellular matrix in morphogenesis. Curr Opin Biotechnol, 2003, 14(5), 526 doi: 10.1016/j.copbio.2003.08.002[42] Xia Y, Whitesides G M. Soft lithography. Ann Rev Mater Sci, 1998, 28(1), 153 doi: 10.1146/annurev.matsci.28.1.153[43] Qin D, Xia Y, Whitesides G M. Soft lithography for micro-and nanoscale patterning. Nat Protoc, 2010, 5(3), 491 doi: 10.1038/nprot.2009.234[44] Xia Y, Kim E, Zhao X M, et al. Complex optical surfaces formed by replica molding against elastomeric masters. Science, 1996, 273(5273), 347 doi: 10.1126/science.273.5273.347[45] Strale P O, Azioune A, Bugnicourt G, et al. Multiprotein printing by light-induced molecular adsorption. Adv Mater, 2016, 28(10), 2024 doi: 10.1002/adma.201504154[46] Chen C S, Mrksich M, Huang S, et al. Geometric control of cell life and death. Science, 1997, 276(5317), 1425 doi: 10.1126/science.276.5317.1425[47] Parsons J T, Horwitz A R, Schwartz M A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol, 2010, 11(9), 633 doi: 10.1038/nrm2957[48] Kanchanawong P, Shtengel G, Pasapera A M, et al. Nanoscale architecture of integrin-based cell adhesions. Nature, 2010, 468(7323), 580 doi: 10.1038/nature09621[49] Wang N, Butler J P, Ingber D E. Mechanotransduction across the cell surface and through the cytoskeleton. Science, 1993, 260(5111), 1124 doi: 10.1126/science.7684161[50] Balaban N Q, Schwarz U S, Riveline D, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol, 2001, 3(5), 466 doi: 10.1038/35074532[51] Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science, 2013, 340(6135), 991 doi: 10.1126/science.1231041[52] Fink J, Carpi N, Betz T, et al. External forces control mitotic spindle positioning. Nat Cell Biol, 2011, 13(7), 771 doi: 10.1038/ncb2269[53] Discher D E, Janmey P, Wang Y L. Tissue cells feel and respond to the stiffness of their substrate. Science, 2005, 310(5751), 1139 doi: 10.1126/science.1116995[54] Du Roure O, Saez A, Buguin A, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA, 2005, 102(7), 2390 doi: 10.1073/pnas.0408482102[55] Shao Y, Fu J. Integrated micro/nanoengineered functional biomaterials for cell mechanics and mechanobiology: a materials perspective. Adv Mater, 2014, 26(10), 1494 doi: 10.1002/adma.201304431[56] Pelham R J, Wang Y L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA, 1997, 94(25), 13661 doi: 10.1073/pnas.94.25.13661[57] Sarangi B R, Gupta M, Doss B L, et al. Coordination between intra-and extracellular forces regulates focal adhesion dynamics. Nano Lett, 2017, 17(1), 399 doi: 10.1021/acs.nanolett.6b04364[58] Mitrossilis D, Fouchard J, Guiroy A, et al. Single-cell response to stiffness exhibits muscle-like behavior. Proc Natl Acad Sci USA, 2009, 106(43), 18243 doi: 10.1073/pnas.0903994106[59] Sunyer R, Conte V, Escribano J, et al. Collective cell durotaxis emerges from long-range intercellular force transmission. Science, 2016, 353(6304), 1157 doi: 10.1126/science.aaf7119[60] Paszek M J, Zahir N, Johnson K R, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell, 2005, 8(3), 241 doi: 10.1016/j.ccr.2005.08.010[61] Levental K R, Yu H, Kass L, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell, 2009, 139(5), 891 doi: 10.1016/j.cell.2009.10.027[62] Weiz S M, Medina-Sánchez M, Schmidt O G. Microsystems for single-cell analysis. Adv Biosyst, 2018, 2(2), 1700193 doi: 10.1002/adbi.201700193[63] Ghibaudo M, Trichet L, Le Digabel J, et al. Substrate topography induces a crossover from 2D to 3D behavior in fibroblast migration. Biophys J, 2009, 97(1), 357 doi: 10.1016/j.bpj.2009.04.024[64] Clark P, Connolly P, Curtis A S, et al. Topographical control of cell behaviour. I. Simple step cues. Development, 1987, 99(3), 439[65] Yim E K, Reano R M, Pang S W, et al. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials, 2005, 26(26), 5405 doi: 10.1016/j.biomaterials.2005.01.058[66] Whitesides G M. The origins and the future of microfluidics. Nature, 2006, 442(7101), 368 doi: 10.1038/nature05058[67] Douville N J, Zamankhan P, Tung Y C, et al. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab Chip, 2011, 11(4), 609 doi: 10.1039/C0LC00251H[68] Rahimzadeh J, Meng F, Sachs F, et al. Real-time observation of flow-induced cytoskeletal stress in living cells. Am J Physiol-Cell Physiol, 2011, 301(3), C646 doi: 10.1152/ajpcell.00099.2011[69] Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem, 2010, 82(24), 9983 doi: 10.1021/ac100806x[70] Praetorius H A, Spring K R. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol, 2001, 184(1), 71 doi: 10.1007/s00232-001-0075-4[71] Praetorius H A, Spring K R. Removal of the MDCK cell primary cilium abolishes flow sensing. J Membr Biol, 2003, 191(1), 69 doi: 10.1007/s00232-002-1042-4[72] Jang K J, Suh K Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip, 2010, 10(1), 36 doi: 10.1039/B907515A[73] Jang K J, Cho H S, Kang D H, et al. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr Biol, 2011, 3(2), 134 doi: 10.1039/C0IB00018C[74] Jang K J, Mehr A P, Hamilton G A, et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol, 2013, 5(9), 1119 doi: 10.1039/c3ib40049b[75] Baeyens N, Bandyopadhyay C, Coon B G, et al. Endothelial fluid shear stress sensing in vascular health and disease. J Clin Invest, 2016, 126(3), 821 doi: 10.1172/JCI83083[76] Kawashima S, Yokoyama M. Dysfunction of endothelial nitric oxide synthase and atherosclerosis. Arterioscler, Thromb, Vasc Biol, 2004, 24(6), 998 doi: 10.1161/01.ATV.0000125114.88079.96[77] Baeyens N, Mulligan-Kehoe M J, Corti F, et al. Syndecan 4 is required for endothelial alignment in flow and atheroprotective signaling. Proc Natl Acad Sci USA, 2014, 111(48), 17308 doi: 10.1073/pnas.1413725111[78] Conway D E, Breckenridge M T, Hinde E, et al. Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol, 2013, 23(11), 1024 doi: 10.1016/j.cub.2013.04.049[79] Steward R Jr, Tambe D, Hardin C C, et al. Fluid shear, intercellular stress, and endothelial cell alignment. Am J Physiol-Cell Physiol, 2015, 308(8), C657 doi: 10.1152/ajpcell.00363.2014[80] Hur S S, Del Alamo J C, Park J S, et al. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc Natl Acad Sci USA, 2012, 109(28), 11110 doi: 10.1073/pnas.1207326109[81] Ohta S, Inasawa S, Yamaguchi Y. Alignment of vascular endothelial cells as a collective response to shear flow. J Phys D, 2015, 48(24), 245401 doi: 10.1088/0022-3727/48/24/245401[82] Lee H J, Diaz M F, Price K M, et al. Fluid shear stress activates YAP1 to promote cancer cell motility. Nat Commun, 2017, 8, 14122 doi: 10.1038/ncomms14122[83] Polacheck W J, Charest J L, Kamm R D. Interstitial flow influences direction of tumor cell migration through competing mechanisms. Proc Natl Acad Sci USA, 2011, 108(27), 11115 doi: 10.1073/pnas.1103581108[84] Piotrowski-Daspit A S, Tien J, Nelson C M. Interstitial fluid pressure regulates collective invasion in engineered human breast tumors via Snail, vimentin, and E-cadherin. Integr Biol, 2016, 8(3), 319 doi: 10.1039/c5ib00282f[85] Bhatia S N, Ingber D E. Microfluidic organs-on-chips. Nat Biotechnol, 2014, 32(8), 760 doi: 10.1038/nbt.2989[86] Li L, Eyckmans J, Chen C S. Designer biomaterials for mechanobiology. Nat Mater, 2017, 16(12), 1164 doi: 10.1038/nmat5049 -
Proportional views






 DownLoad:
DownLoad: